Difference between revisions of "Ontology for Parasite Life Cycle"
| Line 384: | Line 384: | ||
<ul> | <ul> | ||
<li>[[Leishmania]]</li> | <li>[[Leishmania]]</li> | ||
| − | <li>[[Leishmania | + | <li>[[Leishmania major]]</li> |
<li>[[Leishmania major Amastigote]]</li> | <li>[[Leishmania major Amastigote]]</li> | ||
<li>[[Leishmania major Amastigote Stage]]</li> | <li>[[Leishmania major Amastigote Stage]]</li> | ||
Revision as of 16:15, 4 February 2011
| What is Parasite Lifecycle Ontology (PLO)? |
|
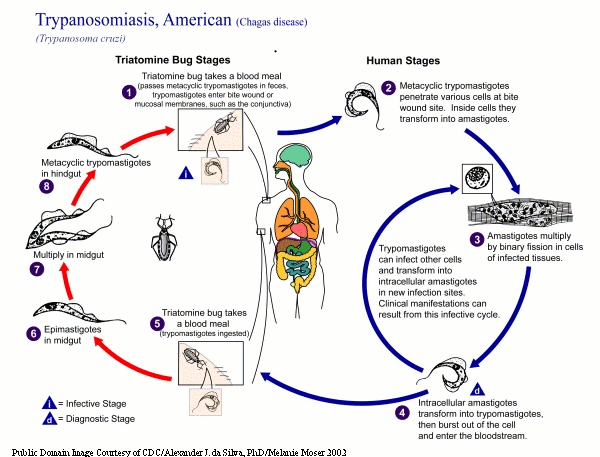
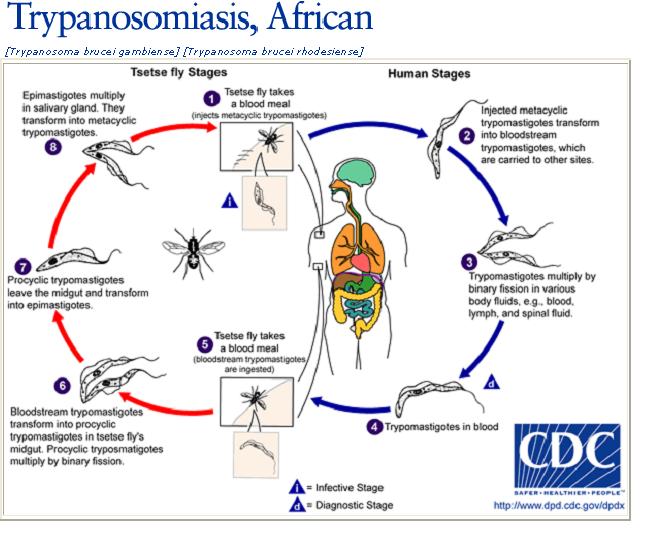
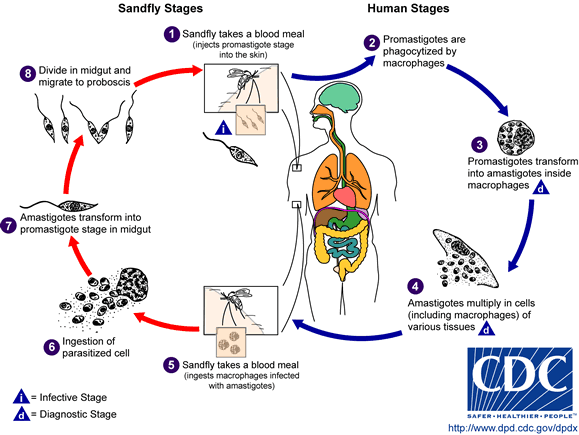
PLO models the life cycle stage details of T.cruzi and two related kinetoplastids, Trypanosoma brucei and Leishmania major. In addition, the ontology also models necessary contextual details such as host information, vector information, strain and anatomical location. All the entities in the ontology are linked to each other by explicitly modeled named relationships that will enable software applications to accurately interpret data annotated with this ontology. For example, “Trypanosoma_cruzi→has_vector_organism→triatominae”. The PLO is modeled in the W3C Web Ontology Language (OWL) standard and currently has 41 classes and 5 properties with a description logic expressivity of ALU. PLO (ver 0.1) is released through NCBO’s BioPortal repository for public use. |
|
|
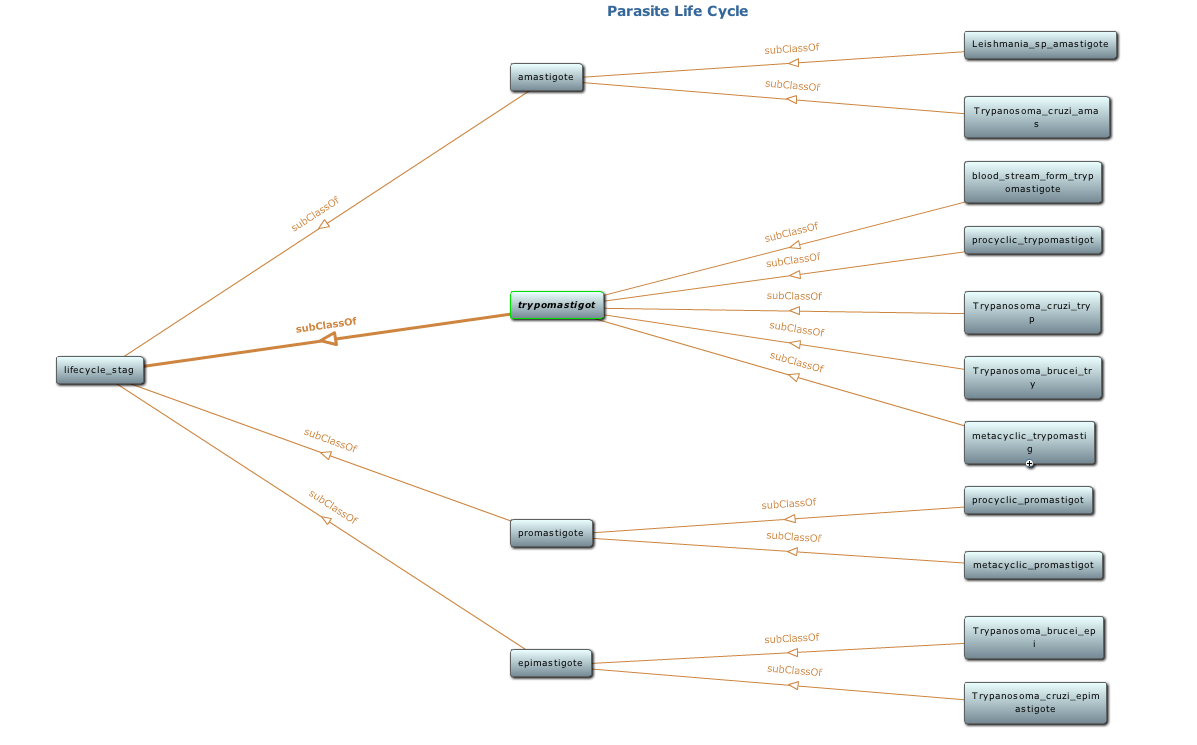 Snapshot of class hierarchy of the parasite life cycle ontology using Protege toolkit. Detail of this ontology is available at NCBO BioPortal |
| Organisms in PLO | ||||||
|
Here is brief description and life-cycles of the organisms that are covered in PLO.
|
| Feedback/Comments |
|
Please add feedback/comments in Discussion Section on the top of this wiki page. |