Parasite Experiment ontology
| What is Parasite Experiment Ontology (PEO)? |
|
The ontology comprehensively models the processes, instruments, parameters, and sample details that will be used to annotate experimental results with “provenance” metadata (derivation history of results). For example, given a drug selected sample (a sample that has been grown under selective drug pressure. source: Parasite Experiment ontology), details about the drug, drug concentration, the duration for the drug selection process, and the transfected sample used as input to the drug selection process are modeled in the ontology. This extensive provenance metadata modeled in this ontology will enable publication of results in journals, conferences with details of the method used to arrive at the result. PEO has 110 classes and 27 properties with a logic expressivity of ALCHQ(D) and incorporates concepts defined in Parasite Life Cycle ontology (PLO) to maximize reuse of existing ontological resources. The new version of PEO (ver 0.2) was released for listing and public use at NCBO’s BioPortal in December 2009. |
|
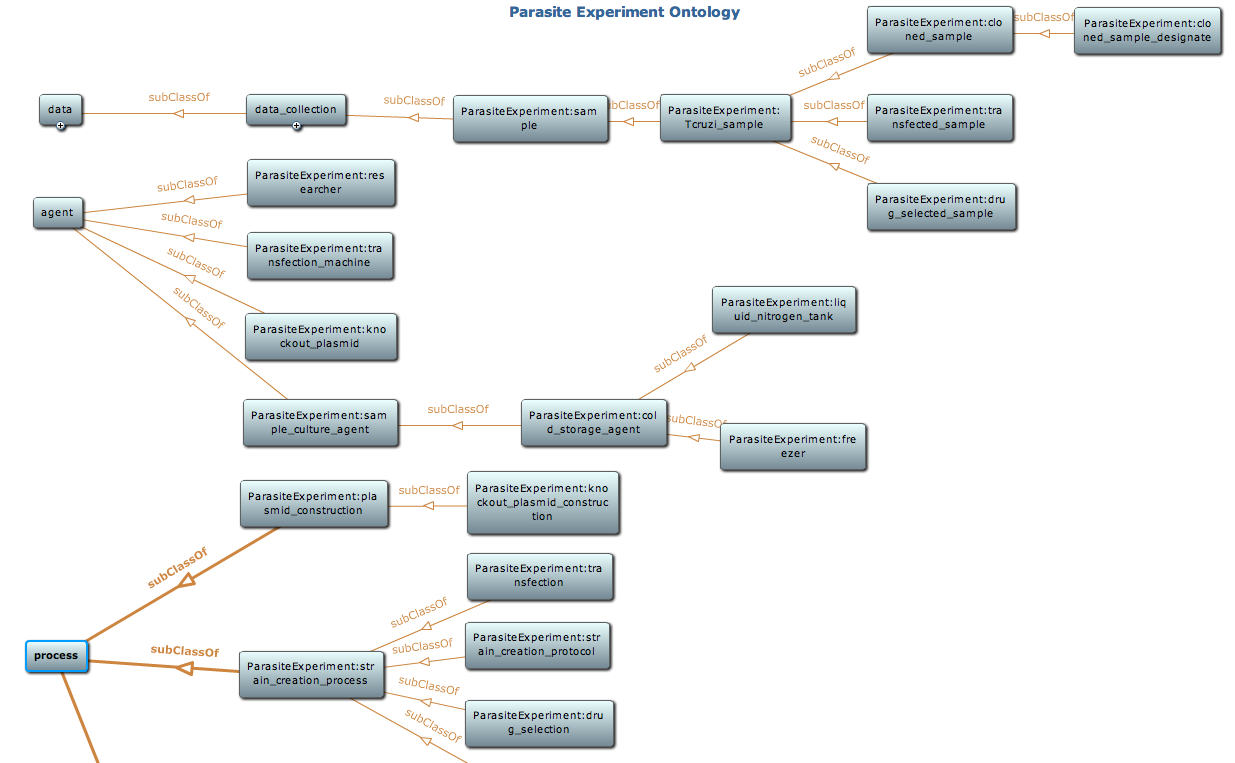 Snapshot of class hierarchy of the parasite experiment ontology using Protege toolkit. Detail of this ontology is available at NCBO BioPortal |
| Alignment of PEO and OBI |
|
These two ontologies present an interesting but different scenario from the OAEI anatomy subtrack. The PEO is being collaboratively developed as part of an NIH funded project to develop and deploy an ontology-driven semantic problem-solving environment for parasite research. The PEO models experiment details and provenance information. It is an application-oriented and more specific domain ontology than OBI, which describes biomedical investigations. In contrast, the OAEI anatomy subtrack ontologies have many common entities since both ontologies describe the same domain (i.e. anatomy) for different species (mouse vs. human). Mapping more specific ontologies such as PEO to a more general ontology such as OBI is an important step towards helping establish a common point of reference that can serve to foster cooperation and interoperability among researchers. Given the explosion of metadata available on the web, our case study to align two related ontologies in the biomedical field has the potential to impact not just the biomedical field but also any research fields that take advantage of semantic web technologies. |
Mappings between PEO and OBI submitted to NCBO BioPortal:
| Feedback/Comments |
|
Please add feedback/comments in Discussion Section on the top of this wiki page. |