Difference between revisions of "Trykipedia: T.cruzi Semantics and Services enabled Problem Solving Environment"
Cotent-admin (Talk | contribs) m (31 revisions) |
|||
| (22 intermediate revisions by 2 users not shown) | |||
| Line 25: | Line 25: | ||
/ref | /ref | ||
) and proteome analyses are also available. These dynamic datasets, addressing different but logically related aspects, are distributed over multiple databases that undergo frequent additions and curation. | ) and proteome analyses are also available. These dynamic datasets, addressing different but logically related aspects, are distributed over multiple databases that undergo frequent additions and curation. | ||
| − | |||
| − | |||
| − | [[Image:Trypanosoma_cruzi.gif|right| | + | |
| + | [[Image:Trypanosoma_cruzi.gif|right|T.cruzi Research Picture]] | ||
| + | |||
== Semantics and Services enabled Problem Solving Environment for Tcruzi Project== | == Semantics and Services enabled Problem Solving Environment for Tcruzi Project== | ||
| − | The Semantics and Services enabled Problem Solving Environment for ''T. cruzi'' project aims to utilize state-of-the-art semantic technologies for effective querying of multiple databases through creation of a suite of ontologies modeling multiple aspects of ''T. cruzi'' research domain. Trykipedia is a Wiki-based resource for the ontologies being created as part of this project | + | The Semantics and Services enabled Problem Solving Environment for ''T. cruzi'' project aims to utilize state-of-the-art semantic technologies for effective querying of multiple databases through creation of a suite of ontologies modeling multiple aspects of ''T. cruzi'' research domain. Trykipedia is a Wiki-based resource for the ontologies being created as part of this project. |
br/ | br/ | ||
br/ | br/ | ||
| + | [http://www.knoesis.org/research/semsci/application_domain/sem_life_sci/tcruzi_pse/ Project Page] | ||
| − | == Parasite Life Cycle | + | == Parasite Life Cycle ontology == |
| − | We have completed the development of an ontology to model the life cycle stage details of T.cruzi and two related kinetoplastids, Trypanosoma brucei and Leishmania major. In addition to life cycle stages, the ontology also models necessary contextual details such as host information, vector information, strain and, anatomical location | + | We have completed the development of an ontology to model the life cycle stage details of T.cruzi and two related kinetoplastids, Trypanosoma brucei and Leishmania major. In addition to life cycle stages, the ontology also models necessary contextual details such as host information, vector information, strain and, anatomical location. |
| + | |||
| + | [[Parasite Life Cycle ontology | ontology details]] | ||
== TcruziExperiri ontology == | == TcruziExperiri ontology == | ||
| − | We have also completed the development of the TcruziExperiri ontology that models the transcriptome and gene-knockout phases of T.cruzi experiment protocols. The ontology comprehensively models the processes, instruments, parameters, and sample details | + | We have also completed the development of the TcruziExperiri ontology that models the transcriptome and gene-knockout phases of T.cruzi experiment protocols. The ontology comprehensively models the processes, instruments, parameters, and sample details that will be used to annotate experimental results with “provenance” metadata (derivation history of results). |
| + | [[TcruziExperiri ontology | ontology details]] | ||
| + | br/ | ||
| + | |||
| + | == T.cruzi Resource Articles == | ||
| + | table width=100% | ||
| + | tr valign=top | ||
| + | td | ||
| + | ;A | ||
| + | ul | ||
| + | li [[Accession Number]]/li | ||
| + | li[[Amastigote]]/li | ||
| + | li[[Anatomical Location]] | ||
| + | /li | ||
| + | /ul | ||
| + | |||
| + | ;B | ||
| + | ul | ||
| + | li[[Blood stream form]]/li | ||
| + | li[[Bloodstream]]/li | ||
| + | /ul | ||
| + | |||
| + | ;C | ||
| + | ul | ||
| + | li[[Cell Cloning]]/li | ||
| + | li[[Chagas disease]]/li | ||
| + | li[[Cloned Sample]]/li | ||
| + | /ul | ||
| + | |||
| + | ;D | ||
| + | ul | ||
| + | li[[Dog]]/li | ||
| + | li[[Drug]]/li | ||
| + | li[[Drug Concentration]]/li | ||
| + | li[[Drug Resistant Plasmid]]/li | ||
| + | li[[Drug Selected Sample]]/li | ||
| + | li[[Drug Selection]]/li | ||
| + | /ul | ||
| + | ;E | ||
| + | ul | ||
| + | li[[Epimastigote]]/li | ||
| + | /ul | ||
| + | ;G | ||
| + | ul | ||
| + | li[[Gene Knockout Protocol]]/li | ||
| + | li[[Genome]]/li | ||
| + | /ul | ||
| + | |||
| + | ;H | ||
| + | ul | ||
| + | li[[Hindgut]]/li | ||
| + | li[[Host organism]]/li | ||
| + | li[[Human]]/li | ||
| + | li[[Human African Trypanosomiasis]]/li | ||
| + | li[[Hybridisation]]/li | ||
| + | /ul | ||
| + | ;K | ||
| + | ul | ||
| + | li[[Kinetoplastid]]/li | ||
| + | li[[Knockout Construct Plasmid]]/li | ||
| + | /ul | ||
| + | /td | ||
| + | |||
| + | td | ||
| + | |||
| + | ;K cont. | ||
| + | ul | ||
| + | li[[Knockout Plasmid]]/li | ||
| + | /ul | ||
| + | |||
| + | ;L | ||
| + | ul | ||
| + | li[[Labelling]]/li | ||
| + | li[[Leishmania]]/li | ||
| + | li[[Leishmaniasis]]/li | ||
| + | li[[Lifecycle stage]]/li | ||
| + | li[[Lutzomyia]]/li | ||
| + | /ul | ||
| + | |||
| + | |||
| + | |||
| + | ;M | ||
| + | ul | ||
| + | li[[Macrophage]]/li | ||
| + | li[[Medium]]/li | ||
| + | li[[Metacyclic]]/li | ||
| + | li[[Mice]]/li | ||
| + | li[[Microarray Analysis]]/li | ||
| + | li[[Microarray Plate Reader]]/li | ||
| + | li[[Midgut]]/li | ||
| + | /ul | ||
| + | |||
| + | ;N | ||
| + | ul | ||
| + | li[[New World]]/li | ||
| + | li[[Nucleated Cell]]/li | ||
| + | li[[Nucleic Acid]]/li | ||
| + | /ul | ||
| + | |||
| + | |||
| + | ;O | ||
| + | ul | ||
| + | li[[Old World]]/li | ||
| + | li[[Ontology]]/li | ||
| + | li[[Organism]]/li | ||
| + | /ul | ||
| + | |||
| + | |||
| + | ;P | ||
| + | ul | ||
| + | li[[Parasitic organism]]/li | ||
| + | li[[Phenotype]]/li | ||
| + | li[[Phlebotomus]]/li | ||
| + | li[[Plasmid]]/li | ||
| + | li[[Plasmid Construction]]/li | ||
| + | li[[Polymerase Chain Reaction]]/li | ||
| + | /ul | ||
| + | |||
| + | /td | ||
| + | |||
| + | td | ||
| + | |||
| + | ;P cont. | ||
| + | ul | ||
| + | li[[Prehybridisation]]/li | ||
| + | li[[Procyclic]]/li | ||
| + | li[[Promastigote]]/li | ||
| + | li[[Purity]]/li | ||
| + | /ul | ||
| + | |||
| + | |||
| + | ;S | ||
| + | ul | ||
| + | li[[Salivary Gland]]/li | ||
| + | li[[Sample]]/li | ||
| + | li[[Sand fly]]/li | ||
| + | li[[Sequence Extraction]]/li | ||
| + | li[[Signal Intensity]]/li | ||
| + | li[[Sleeping sickness]]/li | ||
| + | li[[Strains]]/li | ||
| + | /ul | ||
| + | |||
| + | |||
| + | ;T | ||
| + | ul | ||
| + | li[[Tcruzi Sample]]/li | ||
| + | li[[Transcriptome Protocol]]/li | ||
| + | li[[Transfected Sample]]/li | ||
| + | li[[Transfection]]/li | ||
| + | li[[Transfection Buffer]]/li | ||
| + | li[[Transfection Machine]]/li | ||
| + | li[[Triatomine]]/li | ||
| + | li[[Trypanosoma brucei]]/li | ||
| + | li[[Trypanosoma cruzi]]/li | ||
| + | li[[Trypomastigote]]/li | ||
| + | li[[Tsetse fly]]/li | ||
| + | /ul | ||
| + | |||
| + | ;V | ||
| + | ul | ||
| + | li[[Vector organism]]/li | ||
| + | /ul | ||
| + | |||
| + | |||
| + | /td | ||
| + | |||
| + | |||
| + | /tr | ||
| + | /table | ||
| + | |||
| + | br/ | ||
| + | |||
| + | == Publications == | ||
| + | 1. Prashant Doshi, Ravikanth Kolli, Christopher Thomas, Inexact matching of ontology graphs using expectation-maximization, Journal of Web Semantics: Science, Services and Agents on the World Wide Web, In Press 29 January 2009, ISSN 1570-8268, DOI: 10.1016/j.websem.2008.12.001. | ||
| + | br/ | ||
| + | 2. Amit Sheth , Rick Tarleton , Prashant Doshi , Mark Musen , Natasha Noy , Satya S. Sahoo , D. B. Weatherly , Pablo N. Mendes, “Collaborative RO1 with NCBO Semantics and Services Enabled Problem Solving Environment For Trypanosoma Cruzi”, NCBC-AHM08, poster [http://www.knoesis.org/library/resource.html?id=00350 link] | ||
| + | br/ | ||
| + | 3. Satya S. Sahoo, Roger S. Barga, Jonathan Goldstein, Amit Sheth, Provenance Algebra and Materialized View-based Provenance Management, Microsoft Research Technical Report, (MSR-TR-2008-170) November 2008 [http://knoesis.wright.edu/library/resource.php?id=00354 link] | ||
| + | br/ | ||
br/ | br/ | ||
References:references/ | References:references/ | ||
Latest revision as of 14:54, 11 March 2009
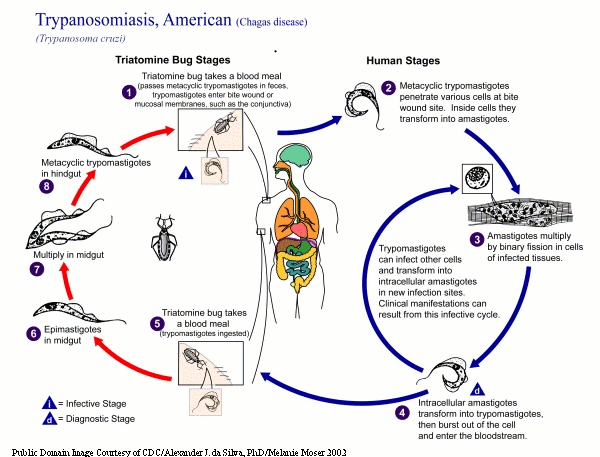
Trypanosoma cruzi is a protozoan parasite and a relative of other human pathogens that cause African sleeping sickness and leishmaniasis. Approximately 18 million people in Latin America are infected with this parasite, and as many as 40% of these are predicted to eventually suffer from Chagas disease, which is the leading cause of heart disease and sudden death in middle-aged adults in the region. Currently, there is a single manufactured drug for treatment of the infection; but this drug has severe side effects and is also carcinogenic. Diagnosis of the infection is poor and there are no vaccines for prevention of the infection. Moreover, T. cruzi presents a level of complexity that, though not unique, is substantial. br/ br/ Research in many biological systems, particularly those involving infectious agents, continues to experience an enormous increase in information with the sequencing of genomes, the use of expression profiling, and the completion of proteomic analyses. For example, annotated genomes and comparative analyses of two T. cruzi related kinetoplastids, Trypanosoma brucei ref Berriman M, G.E., Hertz-Fowler C, Blandin G, Renauld H, Bartholomeu DC, Lennard NJ, Caler E, Hamlin NE, Haas B, Bohme U, Hannick L, Aslett MA, Shallom J, Marcello L, Hou L, Wickstead B, Alsmark UC, Arrowsmith C, Atkin RJ, Barron AJ, Bringaud F, Brooks K, Carrington M, Cherevach I, Chillingworth TJ, Churcher C, Clark LN, Corton CH, Cronin A, Davies RM, Doggett J, Djikeng A, Feldblyum T, Field MC, Fraser A, Goodhead I, Hance Z, Harper D, Harris BR, Hauser H, Hostetler J, Ivens A, Jagels K, Johnson D, Johnson J, Jones K, Kerhornou AX, Koo H, Larke N, Landfear S, Larkin C, Leech V, Line A, Lord A, Macleod A, Mooney PJ, Moule S, Martin DM, Morgan GW, Mungall K, Norbertczak H, Ormond D, Pai G, Peacock CS, Peterson J, Quail MA, Rabbinowitsch E, Rajandream MA, Reitter C, Salzberg SL, Sanders M, Schobel S, Sharp S, Simmonds M, Simpson AJ, Tallon L, Turner CM, Tait A, Tivey AR, Van Aken S, Walker D, Wanless D, Wang S, White B, White O, Whitehead S, Woodward J, Wortman J, Adams MD, Embley TM, Gull K, Ullu E, Barry JD, Fairlamb AH, Opperdoes F, Barrell BG, Donelson JE, Hall N, Fraser CM, Melville SE, El-Sayed NM., The genome of the African trypanosome Trypanosoma brucei. Science, 2005 Jul 15. 309(5733): p. 416-22. /ref and Leishmania major ref Ivens AC, P.C., Worthey EA, Murphy L, Aggarwal G, Berriman M, Sisk E, Rajandream MA, Adlem E, Aert R, Anupama A, Apostolou Z, Attipoe P, Bason N, Bauser C, Beck A, Beverley SM, Bianchettin G, Borzym K, Bothe G, Bruschi CV, Collins M, Cadag E, Ciarloni L, Clayton C, Coulson RM, Cronin A, Cruz AK, Davies RM, De Gaudenzi J, Dobson DE, Duesterhoeft A, Fazelina G, Fosker N, Frasch AC, Fraser A, Fuchs M, Gabel C, Goble A, Goffeau A, Harris D, Hertz-Fowler C, Hilbert H, Horn D, Huang Y, Klages S, Knights A, Kube M, Larke N, Litvin L, Lord A, Louie T, Marra M, Masuy D, Matthews K, Michaeli S, Mottram JC, Muller-Auer S, Munden H, Nelson S, Norbertczak H, Oliver K, O'neil S, Pentony M, Pohl TM, Price C, Purnelle B, Quail MA, Rabbinowitsch E, Reinhardt R, Rieger M, Rinta J, Robben J, Robertson L, Ruiz JC, Rutter S, Saunders D, Schafer M, Schein J, Schwartz DC, Seeger K, Seyler A, Sharp S, Shin H, Sivam D, Squares R, Squares S, Tosato V, Vogt C, Volckaert G, Wambutt R, Warren T, Wedler H, Woodward J, Zhou S, Zimmermann W, Smith DF, Blackwell JM, Stuart KD, Barrell B, Myler PJ., The genome of the kinetoplastid parasite, Leishmania major. Science, 2005 Jul 15. 309(5733): p. 436-42. /ref , and a whole organism (all life-cycle stages) proteome analysis of T. cruzi have been published ref Atwood, J.A., et al., The Trypanosoma cruzi Proteome, in Science 2005. p. 473 – 476. /ref . Data from other sequencing projects in T. cruzi, including ESTs ref Agüero F, Z.W., Weatherly DB, Mendes P, Kissinger JC, TcruziDB: an integrated, postgenomics community resource for Trypanosoma cruzi. Nucleic Acids Research, Jan 1 2006. 34(Database issue): p. D428-31. /ref , preexisting and ongoing expression profiling (DNA microarray ref Minning TA, B.J., Garcia GA, McGraw RA, Tarleton RL, Microarray profiling of gene expression during trypomastigote to amastigote transition in Trypanosoma cruzi. Mol Biochem Parasitol., 2003 Sep. 131(1): p. 55-64. /ref ) and proteome analyses are also available. These dynamic datasets, addressing different but logically related aspects, are distributed over multiple databases that undergo frequent additions and curation.
Contents
Semantics and Services enabled Problem Solving Environment for Tcruzi Project
The Semantics and Services enabled Problem Solving Environment for T. cruzi project aims to utilize state-of-the-art semantic technologies for effective querying of multiple databases through creation of a suite of ontologies modeling multiple aspects of T. cruzi research domain. Trykipedia is a Wiki-based resource for the ontologies being created as part of this project. br/ br/
Parasite Life Cycle ontology
We have completed the development of an ontology to model the life cycle stage details of T.cruzi and two related kinetoplastids, Trypanosoma brucei and Leishmania major. In addition to life cycle stages, the ontology also models necessary contextual details such as host information, vector information, strain and, anatomical location.
TcruziExperiri ontology
We have also completed the development of the TcruziExperiri ontology that models the transcriptome and gene-knockout phases of T.cruzi experiment protocols. The ontology comprehensively models the processes, instruments, parameters, and sample details that will be used to annotate experimental results with “provenance” metadata (derivation history of results).
ontology details br/
T.cruzi Resource Articles
table width=100%
tr valign=top
td
- A
ul li Accession Number/li liAmastigote/li liAnatomical Location /li /ul
- B
ul
liBlood stream form/li
liBloodstream/li
/ul
- C
ul
liCell Cloning/li
liChagas disease/li
liCloned Sample/li
/ul
- D
ul
liDog/li
liDrug/li
liDrug Concentration/li
liDrug Resistant Plasmid/li
liDrug Selected Sample/li
liDrug Selection/li
/ul
- E
ul
liEpimastigote/li
/ul
- G
ul
liGene Knockout Protocol/li
liGenome/li
/ul
- H
ul
liHindgut/li
liHost organism/li
liHuman/li
liHuman African Trypanosomiasis/li
liHybridisation/li
/ul
- K
ul
liKinetoplastid/li
liKnockout Construct Plasmid/li
/ul
/td
td
- K cont.
ul
liKnockout Plasmid/li
/ul
- L
ul
liLabelling/li
liLeishmania/li
liLeishmaniasis/li
liLifecycle stage/li
liLutzomyia/li
/ul
- M
ul
liMacrophage/li
liMedium/li
liMetacyclic/li
liMice/li
liMicroarray Analysis/li
liMicroarray Plate Reader/li
liMidgut/li
/ul
- N
ul
liNew World/li
liNucleated Cell/li
liNucleic Acid/li
/ul
- O
ul
liOld World/li
liOntology/li
liOrganism/li
/ul
- P
ul
liParasitic organism/li
liPhenotype/li
liPhlebotomus/li
liPlasmid/li
liPlasmid Construction/li
liPolymerase Chain Reaction/li
/ul
/td
td
- P cont.
ul
liPrehybridisation/li
liProcyclic/li
liPromastigote/li
liPurity/li
/ul
- S
ul
liSalivary Gland/li
liSample/li
liSand fly/li
liSequence Extraction/li
liSignal Intensity/li
liSleeping sickness/li
liStrains/li
/ul
- T
ul
liTcruzi Sample/li
liTranscriptome Protocol/li
liTransfected Sample/li
liTransfection/li
liTransfection Buffer/li
liTransfection Machine/li
liTriatomine/li
liTrypanosoma brucei/li
liTrypanosoma cruzi/li
liTrypomastigote/li
liTsetse fly/li
/ul
- V
ul
liVector organism/li
/ul
/td
/tr
/table
br/
Publications
1. Prashant Doshi, Ravikanth Kolli, Christopher Thomas, Inexact matching of ontology graphs using expectation-maximization, Journal of Web Semantics: Science, Services and Agents on the World Wide Web, In Press 29 January 2009, ISSN 1570-8268, DOI: 10.1016/j.websem.2008.12.001. br/ 2. Amit Sheth , Rick Tarleton , Prashant Doshi , Mark Musen , Natasha Noy , Satya S. Sahoo , D. B. Weatherly , Pablo N. Mendes, “Collaborative RO1 with NCBO Semantics and Services Enabled Problem Solving Environment For Trypanosoma Cruzi”, NCBC-AHM08, poster link br/ 3. Satya S. Sahoo, Roger S. Barga, Jonathan Goldstein, Amit Sheth, Provenance Algebra and Materialized View-based Provenance Management, Microsoft Research Technical Report, (MSR-TR-2008-170) November 2008 link br/ br/
References:references/