Difference between revisions of "Scooner"
| Line 50: | Line 50: | ||
[[File:scooner-arch.jpg|center|750px]] | [[File:scooner-arch.jpg|center|750px]] | ||
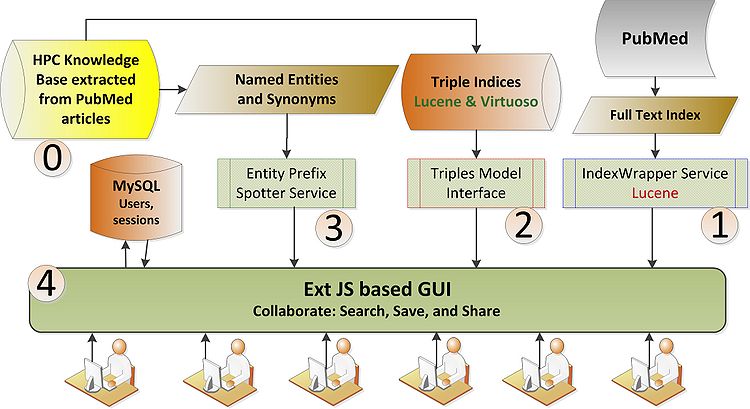
'''0. Knowledge base: ''' We assume the presence of a knowledge base in the form of triples preferably in RDF. Having a good schema for the data really helps with Scooner's functionality. Other formats can also be converted to RDF with some pre-processing. Currently we are hosting two data sets both related to human performance and cognition. One is built at the Knoesis center using NLP and pattern based extraction techniques and is serialized as a Lucene index. The second data set comes from shallow parsing and rule based extraction techniques developed at the NLM and is stored in a Virtuoso triple store. | '''0. Knowledge base: ''' We assume the presence of a knowledge base in the form of triples preferably in RDF. Having a good schema for the data really helps with Scooner's functionality. Other formats can also be converted to RDF with some pre-processing. Currently we are hosting two data sets both related to human performance and cognition. One is built at the Knoesis center using NLP and pattern based extraction techniques and is serialized as a Lucene index. The second data set comes from shallow parsing and rule based extraction techniques developed at the NLM and is stored in a Virtuoso triple store. | ||
| + | |||
| + | '''0. PubMed full text index: ''' | ||
Revision as of 14:14, 20 February 2011
Overview
The limitations of key word based search are well known in the information retrieval field. These are more evident in life sciences where most of the reliable scientific information is spread across biomedical literature in the form textual journal articles. Unlike the Web, these journal articles are devoid of hyper links and multiple key word based searches need to be performed while aggregating and organizing search results that the user finds interesting. This makes literature search a tedious task in life sciences.
Knowledge-based search systems are proposed as an improvement over conventional search and have gained popularity especially given the availability of many expert curated vocabularies and taxonomies in the biomedical domains. The different classes in a given taxonomy are used to provide faceted search over articles that contain the instances of these classes. These taxonomies and other forms of ontologies are mostly static blocks of well accepted consensual knowledge. Also, most of these standard ontologies have a limited number of predicates (or relationship types) such as "part of" and "is a". We believe the search process can benefit from recently published results that are not well known in the research community and also by relationship types that go beyond the taxonomic ones. Scooner is a knowledge-based literature search and exploration system that is built upon this intuition. We are working on providing more powerful knowledge-based search where recently published results are computationally extracted and used a background KB to guide the search process. The key here is that the knowledge-base that guides search is extracted from the same universe of literature that is being explored.
Search Process: In Scooner, search is modeled as an interactive process where, besides a search box for key word input, the points of interaction are based on domain specific assertions (or triples) of the form: subject -> predicate -> object (ex: muscarinic activation -> facilitates -> long-term potentiation). Raw text results are input to a spotter module that annotates them with entities found in the triples used as background knowledge. Clicking on an annotated entity displays all triples where it participates as a subject or object. Clicking on the corresponding object/subject would then bring up articles that potentially contain that triple; in most cases the original abstract from which the triple was extracted is listed in the top 2 or 3 articles. This way the triples can be browsed in the context of the abstracts in which they were found. New implicit knowledge can also be discovered by building trails from individual triples.
Collaborative Extensions: Scooner combines these ideas of triple-based search and exploration with persistent search sessions. Users can create search projects and store their search history including the abstracts they felt important, triples they found useful, and also collaborate with colleagues. The workbench in Scooner facilitates a central aggregation of important abstracts imported for further review. The work bench can be filtered to only show only those abstracts that pertain to a selected set of triples or trails. Additionally, collaborative features were incorporated using which users can create persistent search projects, write comments on abstracts they find relevant, and share the (sub) projects with other users on a public dashboard.
Currently Scooner's KB comes from the human performance and cognition ontology project and the literature explored is the set of all abstracts available via PubMed as of Oct 2010. The knowledge-base is created for the domain of human performance and cognition and is extracted from articles on PubMed published by Aug 2008. Initial evaluations of Scooner by researchers at the AFRL indicate that Scooner does better than NLM's PubMed search tool. For a screencast of Scooner in use, please visit: http://knoesis.wright.edu/library/demos/scooner-demo/
Project Team
Undergraduate Students: Alan Smith, Paul Fultz
Graduate Students: Delroy Cameron, Christopher Thomas, Wenbo Wang
Postdocs: Ramakanth Kavuluru
Faculty: Amit Sheth
Former students who contributed to previous incarnations of Scooner: Pablo Mendes, Cartic Ramakrishnan
Architecture and Components
The following picture shows various components of Scooner
0. Knowledge base: We assume the presence of a knowledge base in the form of triples preferably in RDF. Having a good schema for the data really helps with Scooner's functionality. Other formats can also be converted to RDF with some pre-processing. Currently we are hosting two data sets both related to human performance and cognition. One is built at the Knoesis center using NLP and pattern based extraction techniques and is serialized as a Lucene index. The second data set comes from shallow parsing and rule based extraction techniques developed at the NLM and is stored in a Virtuoso triple store.
0. PubMed full text index: