Parasite Experiment ontology
| What is Parasite Experiment Ontology (PEO)? |
|
The ontology comprehensively models the processes, instruments, parameters, and sample details that will be used to annotate experimental results with “provenance” metadata (derivation history of results). For example, given a drug selected sample (a sample that has been grown under selective drug pressure. source: Parasite Experiment ontology), details about the drug, drug concentration, the duration for the drug selection process, and the transfected sample used as input to the drug selection process are modeled in the ontology. This extensive provenance metadata modeled in this ontology will enable publication of results in journals, conferences with details of the method used to arrive at the result. PEO has 110 classes and 27 properties with a logic expressivity of ALCHQ(D) and incorporates concepts defined in Parasite Life Cycle ontology (PLO) to maximize reuse of existing ontological resources. The new version of PEO (ver 0.2) was released for listing and public use at NCBO’s BioPortal in December 2009. |
|
|
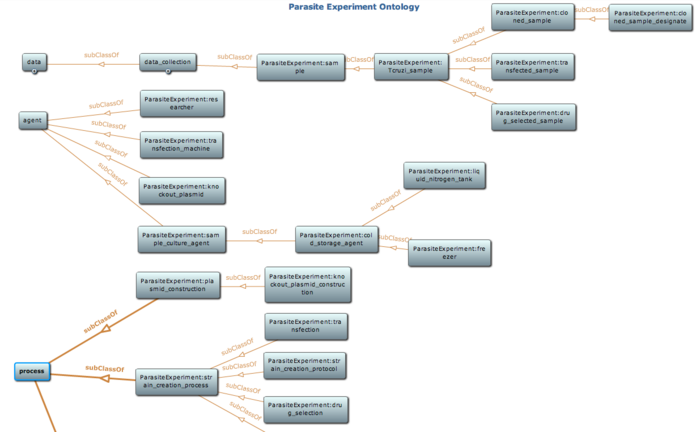 Snapshot of class hierarchy of the parasite experiment ontology using Protege toolkit. Detail of this ontology is available at NCBO BioPortal |
| List of ontology classes |
|
|
Collaborating Institutions
We invite new members to collaborate with us in extending this ontology.
Feedback/Comments
Please add feedback/comments in Discussion Section on the top of this wiki page.