Difference between revisions of "Parasite Experiment ontology"
| (28 intermediate revisions by 3 users not shown) | |||
| Line 14: | Line 14: | ||
{{#tree:id=OwlTree|openlevels=1|root=Owl:Thing| | {{#tree:id=OwlTree|openlevels=1|root=Owl:Thing| | ||
*agent | *agent | ||
| + | **[[Data_Source|data_source]] | ||
**[[Freezer|freezer]] | **[[Freezer|freezer]] | ||
**[[Knockout_Plasmid|knockout_plasmid]] | **[[Knockout_Plasmid|knockout_plasmid]] | ||
| − | |||
**[[Microarray_Plate_Reader|microarray_plate_reader]] | **[[Microarray_Plate_Reader|microarray_plate_reader]] | ||
| + | **[[Researcher|researcher]] | ||
**[[Transfection_Machine|transfection_machine]] | **[[Transfection_Machine|transfection_machine]] | ||
*data | *data | ||
**data_collection | **data_collection | ||
| + | ***[[GO_Term|go_term]] | ||
***[[Peptide_count|peptide_count]] | ***[[Peptide_count|peptide_count]] | ||
***[[Phenotype|phenotype]] | ***[[Phenotype|phenotype]] | ||
***[[Protein_group_number|protein_group_number]] | ***[[Protein_group_number|protein_group_number]] | ||
| − | ***[[sample]] | + | ***[[SAM_result|sam_result]] |
| + | ***[[Sample|sample]] | ||
****[[Tcruzi_Lifecyclestage_Subsample|Tcruzi_lifecyclestage_subsample]] | ****[[Tcruzi_Lifecyclestage_Subsample|Tcruzi_lifecyclestage_subsample]] | ||
****[[Tcruzi_Sample|Tcruzi_sample]] | ****[[Tcruzi_Sample|Tcruzi_sample]] | ||
| Line 49: | Line 52: | ||
****[[Culture_Parameter|culture_parameter]] | ****[[Culture_Parameter|culture_parameter]] | ||
*****[[Temperature_Condition|temperature_condition]] | *****[[Temperature_Condition|temperature_condition]] | ||
| − | ****[[drug]] | + | ****[[Drug|drug]] |
****[[Drug_Attribute|drug_attribute]] | ****[[Drug_Attribute|drug_attribute]] | ||
*****[[Drug_Concentration|drug_concentration]] | *****[[Drug_Concentration|drug_concentration]] | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
****[[Nucleic_Acid|nucleic_acid]] | ****[[Nucleic_Acid|nucleic_acid]] | ||
| + | ****[[Primer|primer]] | ||
| + | *****[[3_Prime_Primer|3_prime_primer]] | ||
| + | ******[[3_Prime_Forward_Primer|3_prime_forward_primer]] | ||
| + | ******[[3_Prime_Reverse_Primer|3_prime_reverse_primer]] | ||
| + | *****[[5_Prime_Primer|5_prime_primer]] | ||
| + | ******[[5_Prime_Forward_Primer|5_prime_forward_primer]] | ||
| + | ******[[5_Prime_Reverse_Primer|5_prime_reverse_primer]] | ||
****[[Purity|purity]] | ****[[Purity|purity]] | ||
****[[Transfection_Buffer|transfection_buffer]] | ****[[Transfection_Buffer|transfection_buffer]] | ||
| − | ****[[ | + | ****[[Host_Organism|host_organism]] |
| − | ****[[ | + | ****[[Lifecycle_Stage|lifecycle_stage]] |
| − | ****[[ | + | *****[[Amastigote|amastigote]] |
| + | ******[[Leishmania_SP_Amastigote|leishmania_sp_amastigote]] | ||
| + | ******[[Trypanosoma_Cruzi_Amastigote|trypanosoma_cruzi_amastigote]] | ||
| + | *****[[Epimastigote|epimastigote]] | ||
| + | ******[[Trypanosoma_Brucei_Epimastigote|trypanosoma_brucei_epimastigote]] | ||
| + | ******[[Trypanosoma_Cruzi_Epimastigote|trypanosoma_cruzi_epimastigote]] | ||
| + | *****[[Promastigote|promastigote]] | ||
| + | ******[[Metacyclic_Promastigote|metacyclic_promastigote]] | ||
| + | ******[[Procyclic_Promastigote|procyclic_promastigote]] | ||
| + | *****[[Trypomastigote|trypomastigote]] | ||
| + | ******[[Trypanosoma_Brucei_Trypomastigote|trypanosoma_brucei_trypomastigote]] | ||
| + | ******[[Trypanosoma_Cruzi_Trypomastigote|trypanosoma_cruzi_trypomastigote]] | ||
| + | ******[[Blood_Stream_Form_Trypomastigote|blood_stream_form_trypomastigote]] | ||
| + | ******[[Metacyclic_Trypomastigote|metacyclic|trypomastigote]] | ||
| + | *******[[Trypanosoma_brucei_Metacyclic_Trypomastigote|trypanosoma_brucei_metacyclic_trypomastigote]] | ||
| + | *******[[Trypanosoma_Cruzi_Metacyclic_Trypomastigote|trypanosoma_cruzi_metacyclic_trypomastigote]] | ||
| + | ******[[Procyclic_Trypomastigote|procyclic_trypomastigote]] | ||
****[[organism]] | ****[[organism]] | ||
****[[Organism_Strain|organism_strain]] | ****[[Organism_Strain|organism_strain]] | ||
| Line 73: | Line 89: | ||
****[[Genome|genome]] | ****[[Genome|genome]] | ||
*****[[Transfected_Genome|transfected_genome]] | *****[[Transfected_Genome|transfected_genome]] | ||
| − | + | ****[[Region|region]] | |
| − | ******[[ | + | *****[[Gene|gene]] |
| − | ******[[ | + | *****[[Intergenic_Region|intergenic_region]] |
| − | ******[[ | + | ******[[3_Prime_Intergenic_Region|3_prime_intergenic_region]] |
| + | *******[[3_Prime_Intergenic_Forward_Region|3_prime_intergenic_forward_region]] | ||
| + | *******[[3_Prime_Intergenic_Reverse_Region|3_prime_intergenic_reverse_region]] | ||
| + | ******[[5_Prime_Intergenic_Region|5_prime_intergenic_region]] | ||
| + | *******[[5_Prime_Intergenic_Forward_Region|5_prime_intergenic_forward_region]] | ||
| + | *******[[5_Prime_Intergenic_Reverse_Region|5_prime_intergenic_reverse_region]] | ||
| + | ****[[Gene_Function|gene_function]] | ||
***spatial_parameter | ***spatial_parameter | ||
****[[Freezer_Location|freezer_location]] | ****[[Freezer_Location|freezer_location]] | ||
| Line 82: | Line 104: | ||
***temporal_parameter | ***temporal_parameter | ||
****[[Date_Time_Description|DateTimeDescription]] | ****[[Date_Time_Description|DateTimeDescription]] | ||
| + | *****[[DateCompleted|dateCompleted]] | ||
| + | *****[[DateOfLastModification|dateOfLastModification]] | ||
| + | *****[[DateStarted|dateStarted]] | ||
*process | *process | ||
**[[Gene_Knockout_Process|gene_knockout_process]] | **[[Gene_Knockout_Process|gene_knockout_process]] | ||
| Line 113: | Line 138: | ||
</tr> | </tr> | ||
</table> | </table> | ||
| + | |||
<!--start section 2 --> | <!--start section 2 --> | ||
| + | |||
| + | {{block | ||
| + | |title= Alignment of PEO and OBI | ||
| + | |title_background=#b0e2ff | ||
| + | |content= These two ontologies present an interesting but different scenario from the OAEI anatomy subtrack. The PEO is being collaboratively developed as part of an NIH funded project to develop and deploy an ontology-driven semantic problem-solving environment for parasite research. The PEO models experiment details and provenance information. It is an application-oriented and more specific domain ontology than OBI, which describes biomedical investigations. In contrast, the OAEI anatomy subtrack ontologies have many common entities since both ontologies describe the same domain (i.e. anatomy) for different species (mouse vs. human). Mapping more specific ontologies such as PEO to a more general ontology such as OBI is an important step towards helping establish a common point of reference that can serve to foster cooperation and interoperability among researchers. Given the explosion of metadata available on the web, our case study to align two related ontologies in the biomedical field has the potential to impact not just the biomedical field but also any research fields that take advantage of semantic web technologies. | ||
| + | }} | ||
| + | |||
| + | ==Mappings between PEO and OBI submitted to NCBO BioPortal:== | ||
| + | |||
| + | {| border="1" cellpadding="5" | ||
| + | |- bgcolor=lightgrey | ||
| + | ! #No !!PEO !! Term!! OBI !! Term !! Relation !! Link | ||
| + | |- align="left" | ||
| + | | 1 ||http://knoesis.wright.edu/provenir/provenir.owl#agent || agent ||http://purl.obolibrary.org/obo/OBI_0000086 || reagent role|| http://www.w3.org/2004/02/skos/core#closeMatch ||http://bioportal.bioontology.org/visualize/42093/?conceptid=agent&jump_to_nav=true#mappings | ||
| + | |- align="left" | ||
| + | | 2||http://knoesis.wright.edu/ParasiteExperiment.owl#standard_deviation || standard_deviation || http://purl.obolibrary.org/obo/OBI_0200121 || standard deviation calculation || http://www.w3.org/2004/02/skos/core#closeMatch || http://bioportal.bioontology.org/visualize/42093/?conceptid=ParasiteExperiment%3Astandard_deviation&jump_to_nav=true#mappings | ||
| + | |- align="left" | ||
| + | | 3||http://ncicb.nci.nih.gov/xml/owl/EVS/Thesaurus.owl#Microarray_Analysis || Microarray_Analysis || http://purl.obolibrary.org/obo/OBI_0400147 || microarray || http://www.w3.org/2004/02/skos/core#closeMatch || http://bioportal.bioontology.org/visualize/42093/?conceptid=Thesaurus%3AMicroarray_Analysis&jump_to_nav=true#mappings | ||
| + | |- align="left" | ||
| + | |4||http://knoesis.wright.edu/ParasiteExperiment.owl#proteome_analysis || proteome_analysis || http://purl.obolibrary.org/obo/OBI_0000615 || protein expression profiling || http://www.w3.org/2004/02/skos/core#closeMatch|| http://bioportal.bioontology.org/visualize/42093/?conceptid=ParasiteExperiment%3Aproteome_analysis#mappings | ||
| + | |- align="left" | ||
| + | | 5||http://purl.org/obo/owl/sequence#region || region || http://purl.org/obo/owl/SO#SO_0000001 || region || http://www.w3.org/2004/02/skos/core#exactMatch|| http://bioportal.bioontology.org/visualize/42093/?conceptid=sequence%3Aregion&jump_to_nav=true#mappings | ||
| + | |- align="left" | ||
| + | |6||http://knoesis.wright.edu/provenir/provenir.owl#process || process || http://www.ifomis.org/bfo/1.1/span#ProcessualEntity || ProcessualEntity ||http://www.w3.org/2004/02/skos/core#exactMatch ||http://bioportal.bioontology.org/visualize/42093/?conceptid=process&jump_to_nav=true#mappings | ||
| + | |- align="left" | ||
| + | |7||http://purl.org/obo/owl/sequence#plasmid || plasmid ||http://purl.obolibrary.org/obo/OBI_0000430 ||plasmid ||http://www.w3.org/2004/02/skos/core#exactMatch ||http://bioportal.bioontology.org/visualize/42093/?conceptid=sequence%3Aplasmid&jump_to_nav=true#mappings | ||
| + | |- align="left" | ||
| + | |8||http://knoesis.wright.edu/ParasiteExperiment.owl#drug ||drug || http://purl.obolibrary.org/obo/OBI_0000040 || drug role || http://www.w3.org/2004/02/skos/core#exactMatch||http://bioportal.bioontology.org/visualize/42093/?conceptid=ParasiteExperiment%3Adrug&jump_to_nav=true#mappings | ||
| + | |- align="left" | ||
| + | |9|| http://knoesis.wright.edu/ParasiteExperiment.owl#nucleic_acid || nucleic_acid || http://purl.org/obo/owl/CHEBI#CHEBI_33696 || nucleic acid || http://www.w3.org/2004/02/skos/core#exactMatch||http://bioportal.bioontology.org/visualize/42093/?conceptid=ParasiteExperiment%3Anucleic_acid&jump_to_nav=true#mappings | ||
| + | |- align="left" | ||
| + | |10|| http://knoesis.wright.edu/ParasiteExperiment.owl#culture_medium ||culture_medium || http://purl.obolibrary.org/obo/OBI_0000079 || culture medium role || http://www.w3.org/2004/02/skos/core#exactMatch||http://bioportal.bioontology.org/visualize/42093/?conceptid=ParasiteExperiment%3Aculture_medium&jump_to_nav=true#mappings | ||
| + | |- align="left" | ||
| + | | 11||http://knoesis.wright.edu/ParasiteExperiment.owl#transfection || transfection|| http://purl.obolibrary.org/obo/OBI_0600060 || DNA transfection || http://www.w3.org/2004/02/skos/core#exactMatch||http://bioportal.bioontology.org/visualize/42093/?conceptid=ParasiteExperiment%3Atransfection&jump_to_nav=true#mappings | ||
| + | |- align="left" | ||
| + | |12||http://paige.ctegd.uga.edu/ParasiteLifecycle.owl#organism || organism || http://purl.obolibrary.org/obo/OBI_0100026 || organism ||http://www.w3.org/2004/02/skos/core#exactMatch ||http://bioportal.bioontology.org/visualize/42093/?conceptid=ParasiteLifecycle%3Aorganism | ||
| + | |- align="left" | ||
| + | |13|| http://knoesis.wright.edu/ParasiteExperiment.owl#polymerase_chain_reaction|| polymerase_chain_reaction || http://purl.obolibrary.org/obo/OBI_0000415 || polymerase chain reaction ||http://www.w3.org/2004/02/skos/core#exactMatch ||http://bioportal.bioontology.org/visualize/42093/?conceptid=ParasiteExperiment%3Apolymerase_chain_reaction&jump_to_nav=true#mappings | ||
| + | |- align="left" | ||
| + | | 14||http://paige.ctegd.uga.edu/ParasiteLifecycle.owl#lifecycle_stage || lifecycle_stage || hhttp://purl.org/obo/owl/UBERON#UBERON_0000105 || life cycle stage || http://www.w3.org/2004/02/skos/core#exactMatch||http://bioportal.bioontology.org/visualize/42093/?conceptid=ParasiteLifecycle%3Alifecycle_stage&jump_to_nav=true#mappings | ||
| + | |- align="left" | ||
| + | | 15||http://knoesis.wright.edu/ParasiteExperiment.owl#researcher ||researcher ||http://purl.obolibrary.org/obo/OBI_0000116 || worker role ||http://www.w3.org/2004/02/skos/core#exactMatch || http://bioportal.bioontology.org/visualize/44899/?conceptid=obi%3AOBI_0000116#mappings | ||
| + | |- align="left" | ||
| + | | 16|| http://knoesis.wright.edu/ParasiteExperiment.owl#signal_intensity||signal_intensity || http://purl.obolibrary.org/obo/OBI_0000010 || fluorescent reporter intensity || http://www.w3.org/2004/02/skos/core#exactMatch||http://bioportal.bioontology.org/visualize/42093/?conceptid=ParasiteExperiment%3Asignal_intensity&jump_to_nav=true#mappings | ||
| + | |- align="left" | ||
| + | | 17||http://knoesis.wright.edu/ParasiteExperiment.owl#temperature_condition || temperature_condition || http://purl.org/obo/owl/PATO#PATO_0000146 || temperature || http://www.w3.org/2004/02/skos/core#exactMatch||http://bioportal.bioontology.org/visualize/42093/?conceptid=ParasiteExperiment%3Atemperature_condition&jump_to_nav=true#mappings | ||
| + | |- align="left" | ||
| + | | 18||http://knoesis.wright.edu/ParasiteExperiment.owl#purity || purity || http://purl.org/obo/owl/PATO#PATO_0001339 || biomaterial purity || http://www.w3.org/2004/02/skos/core#exactMatch||http://bioportal.bioontology.org/visualize/42093/?conceptid=ParasiteExperiment%3Apurity&jump_to_nav=true#mappings | ||
| + | |- align="left" | ||
| + | | 19||http://knoesis.wright.edu/ParasiteExperiment.owl#sample ||sample || http://purl.obolibrary.org/obo/OBI_0100051|| specimen || http://www.w3.org/2004/02/skos/core#exactMatchhttp://www.w3.org/2004/02/skos/core#exactMatch||http://bioportal.bioontology.org/visualize/42093/?conceptid=ParasiteExperiment%3Asample&jump_to_nav=true#mappings | ||
| + | |- align="left" | ||
| + | |20|| http://knoesis.wright.edu/ParasiteExperiment.owl#microarray_plate_reader || microarray_plate_reader || http://purl.obolibrary.org/obo/OBI_0400104 || array scanner|| http://www.w3.org/2004/02/skos/core#exactMatch | ||
| + | ||http://bioportal.bioontology.org/visualize/42093/?conceptid=ParasiteExperiment%3Amicroarray_plate_reader&jump_to_nav=true#mappings | ||
| + | |} | ||
| + | |||
| + | |||
| + | |||
| + | <!--end section 2 --> | ||
| + | |||
| + | <!--start section 3 --> | ||
{{block | {{block | ||
| Line 122: | Line 208: | ||
<tr valign="top"> | <tr valign="top"> | ||
<td> | <td> | ||
| + | <ul> | ||
| + | <li>[[3 Prime Forward Primer]]</li> | ||
| + | <li>[[3 Prime Intergenic Forward Region]]</li> | ||
| + | <li>[[3 Prime Intergenic Region]]</li> | ||
| + | <li>[[3 Prime Intergenic Reverse Region]]</li> | ||
| + | <li>[[3 Prime Primer]]</li> | ||
| + | <li>[[3 Prime Reverse Primer]]</li> | ||
| + | <li>[[5 Prime Forward Primer]]</li> | ||
| + | <li>[[5 Prime Intergenic Forward Region]]</li> | ||
| + | <li>[[5 Prime Intergenic Region]]</li> | ||
| + | <li>[[5 Prime Intergenic Reverse Region]]</li> | ||
| + | <li>[[5 Prime Primer]]</li> | ||
| + | <li>[[5 Prime Reverse Primer]]</li> | ||
| + | </ul> | ||
;A | ;A | ||
<ul> | <ul> | ||
| − | + | <li> [[Accession Number]]</li> | |
| − | + | ||
<li> [[Amastigote]]</li> | <li> [[Amastigote]]</li> | ||
</ul> | </ul> | ||
| + | |||
| + | ;B | ||
| + | <ul> | ||
| + | <li>[[Blood Stream Form Trypomastigote]]</li> | ||
| + | </ul> | ||
;C | ;C | ||
<ul> | <ul> | ||
| − | + | <li>[[Cell Cloning]]</li> | |
<li>[[Chromosome]]</li> | <li>[[Chromosome]]</li> | ||
| − | + | <li>[[Cloned Sample]]</li> | |
| − | + | <li>[[Cloned Sample Designate]]</li> | |
<li>[[Culture Medium]]</li> | <li>[[Culture Medium]]</li> | ||
<li>[[Culture Parameter]]</li> | <li>[[Culture Parameter]]</li> | ||
| Line 141: | Line 245: | ||
;D | ;D | ||
<ul> | <ul> | ||
| + | <li>[[Data Source]]</li> | ||
| + | <li>[[Date Completed]]</li> | ||
| + | <li>[[Date of Last Modification]]</li> | ||
| + | <li>[[Date Started]]</li> | ||
<li>[[Date Time Description]]</li> | <li>[[Date Time Description]]</li> | ||
<li>[[Drug]]</li> | <li>[[Drug]]</li> | ||
<li>[[Drug Attribute]]</li> | <li>[[Drug Attribute]]</li> | ||
| − | + | <li>[[Drug Concentration]]</li> | |
| − | + | <li>[[Drug Resistant Plasmid]]</li> | |
| − | + | <li>[[Drug Selected Sample]]</li> | |
| − | + | <li>[[Drug Selection]]</li> | |
</ul> | </ul> | ||
;E | ;E | ||
<ul> | <ul> | ||
<li>[[Epimastigote]]</li> | <li>[[Epimastigote]]</li> | ||
| − | |||
| − | |||
</ul> | </ul> | ||
;F | ;F | ||
<ul> | <ul> | ||
| − | |||
<li>[[Freezer]]</li> | <li>[[Freezer]]</li> | ||
<li>[[Freezer Location]]</li> | <li>[[Freezer Location]]</li> | ||
| Line 168: | Line 273: | ||
<ul> | <ul> | ||
<li>[[Gene]]</li> | <li>[[Gene]]</li> | ||
| − | <li>[[Gene Function]]</li> | + | <li>[[Gene Function]]</li> |
| − | <li>[[Gene Knockout | + | <li>[[Gene Knockout Process]]</li> |
<li>[[Genome]]</li> | <li>[[Genome]]</li> | ||
| + | <li>[[GO Term]]</li> | ||
</ul> | </ul> | ||
;H | ;H | ||
<ul> | <ul> | ||
| − | + | <li>[[Host organism]]</li> | |
| − | + | <li>[[Hybridisation]]</li> | |
</ul> | </ul> | ||
;I | ;I | ||
<ul> | <ul> | ||
| − | + | <li>[[Intergenic Region]]</li> | |
</ul> | </ul> | ||
| Line 187: | Line 293: | ||
<li>[[Kinetoplast Position]]</li> | <li>[[Kinetoplast Position]]</li> | ||
<li>[[Knockout Construct Plasmid]]</li> | <li>[[Knockout Construct Plasmid]]</li> | ||
| − | + | <li>[[Knockout Plasmid]]</li> | |
<li>[[Knockout Plasmid Construction]]</li> | <li>[[Knockout Plasmid Construction]]</li> | ||
<li>[[Knockout Project Protocol]]</li> | <li>[[Knockout Project Protocol]]</li> | ||
| Line 195: | Line 301: | ||
;L | ;L | ||
<ul> | <ul> | ||
| − | + | <li>[[Labelling]]</li> | |
<li>[[Laboratory Experiment Parameter]]</li> | <li>[[Laboratory Experiment Parameter]]</li> | ||
| − | + | <li>[[Leishmania SP Amastigote]]</li> | |
| − | <li>[[Log base2 Ratio]]</li> </ul> | + | <li>[[Lifecycle stage]]</li> |
| + | <li>[[Log base2 Ratio]]</li> | ||
| + | </ul> | ||
</td> | </td> | ||
<td> | <td> | ||
;M | ;M | ||
<ul> | <ul> | ||
| − | + | <li>[[Metacyclic_Promastigote]]</li> | |
| − | + | <li>[[Metacyclic_Trypomastigote]]</li> | |
| − | <li>[[Microarray | + | <li>[[Microarray Analysis]]</li> |
<li>[[Microarray Oligonucleotide]]</li> | <li>[[Microarray Oligonucleotide]]</li> | ||
| − | + | <li>[[Microarray Plate Reader]]</li> | |
</ul> | </ul> | ||
;N | ;N | ||
<ul> | <ul> | ||
| − | + | <li>[[Nucleic Acid]]</li> | |
</ul> | </ul> | ||
| Line 224: | Line 332: | ||
;P | ;P | ||
<ul> | <ul> | ||
| − | + | <li>[[Peptide count]]</li> | |
| − | + | <li>[[Phenotype]]</li> | |
| − | + | <li>[[Plasmid]]</li> | |
| − | + | <li>[[Plasmid Construction]]</li> | |
| − | <li>[[ | + | <li>[[Polymerase Chain Reaction]]</li> |
| − | + | <li>[[Prehybridisation]]</li> | |
| − | + | <li>[[Primer]]</li> | |
| − | + | <li>[[Priority]]</li> | |
| − | <li>[[Probability]]</li> | + | <li>[[Probability]]</li> |
| + | <li>[[Procyclic Promastigote]]</li> | ||
| + | <li>[[Procyclic Trypomastigote]]</li> | ||
<li>[[Promastigote]]</li> | <li>[[Promastigote]]</li> | ||
<li>[[Protein group number]]</li> | <li>[[Protein group number]]</li> | ||
| Line 246: | Line 356: | ||
<ul> | <ul> | ||
<li>[[Region]]</li> | <li>[[Region]]</li> | ||
| − | <li>[[Researcher | + | <li>[[Researcher]]</li> |
| − | + | ||
</ul> | </ul> | ||
;S | ;S | ||
<ul> | <ul> | ||
| − | + | <li>[[SAM Result]]</li> | |
| + | <li>[[Sample]]</li> | ||
<li>[[Sample Characterization]]</li> | <li>[[Sample Characterization]]</li> | ||
<li>[[Scanning]]</li> | <li>[[Scanning]]</li> | ||
<li>[[Sequence Coverage]]</li> | <li>[[Sequence Coverage]]</li> | ||
| − | + | <li>[[Sequence Extraction]]</li> | |
| − | + | <li>[[Signal Intensity]]</li> | |
<li>[[Spectral Count]]</li> | <li>[[Spectral Count]]</li> | ||
<li>[[Stage Purity Determination]]</li> | <li>[[Stage Purity Determination]]</li> | ||
| Line 262: | Line 372: | ||
<li>[[Statistical Measure]]</li> | <li>[[Statistical Measure]]</li> | ||
<li>[[Status]]</li> | <li>[[Status]]</li> | ||
| − | |||
<li>[[Strain Creation Process]]</li> | <li>[[Strain Creation Process]]</li> | ||
<li>[[Strain Creation Protocol]]</li> | <li>[[Strain Creation Protocol]]</li> | ||
| Line 270: | Line 379: | ||
<li>[[Target Region Plasmid]]</li> | <li>[[Target Region Plasmid]]</li> | ||
<li>[[Tcruzi Lifecyclestage Subsample]]</li> | <li>[[Tcruzi Lifecyclestage Subsample]]</li> | ||
| − | + | <li>[[Tcruzi Sample]]</li> | |
| − | + | ||
<li>[[Temperature Condition]]</li> | <li>[[Temperature Condition]]</li> | ||
<li>[[Transcriptome Process]]</li> | <li>[[Transcriptome Process]]</li> | ||
| − | |||
<li>[[Transfected Genome]]</li> | <li>[[Transfected Genome]]</li> | ||
<li>[[Transfected Sample]]</li> | <li>[[Transfected Sample]]</li> | ||
| − | + | <li>[[Transfection]]</li> | |
| − | + | <li>[[Transfection Buffer]]</li> | |
| − | + | <li>[[Transfection Machine]]</li> | |
| + | <li>[[Trypanosoma Brucei Epimastigote]]</li> | ||
| + | <li>[[Trypanosoma Brucei Metacyclic Trypomastigote]]</li> | ||
| + | <li>[[Trypanosoma Brucei Trypomastigote]]</li> | ||
| + | <li>[[Trypanosoma Cruzi Amastigote]]</li> | ||
| + | <li>[[Trypanosoma_Cruzi_Epimastigote]]</li> | ||
| + | <li>[[Trypanosoma Cruzi Metacyclic Trypomastigote]]</li> | ||
| + | <li>[[Trypanosoma Cruzi Trypomastigote]]</li> | ||
<li>[[Trypomastigote]]</li> | <li>[[Trypomastigote]]</li> | ||
</ul> | </ul> | ||
| Line 289: | Line 403: | ||
</table> | </table> | ||
}} | }} | ||
| − | <!--end section | + | <!--end section 3 --> |
| − | <!--start section | + | <!--start section 4 --> |
{{block | {{block | ||
| Line 297: | Line 411: | ||
|content=Please add feedback/comments in [http://wiki.knoesis.org/index.php?title=Talk:TcruziExperiri_ontology&action=edit Discussion Section] on the top of this wiki page. | |content=Please add feedback/comments in [http://wiki.knoesis.org/index.php?title=Talk:TcruziExperiri_ontology&action=edit Discussion Section] on the top of this wiki page. | ||
}} | }} | ||
| − | <!--end section | + | <!--end section 4--> |
Latest revision as of 14:13, 17 March 2011
| What is Parasite Experiment Ontology (PEO)? |
|
The ontology comprehensively models the processes, instruments, parameters, and sample details that will be used to annotate experimental results with “provenance” metadata (derivation history of results). For example, given a drug selected sample (a sample that has been grown under selective drug pressure. source: Parasite Experiment ontology), details about the drug, drug concentration, the duration for the drug selection process, and the transfected sample used as input to the drug selection process are modeled in the ontology. This extensive provenance metadata modeled in this ontology will enable publication of results in journals, conferences with details of the method used to arrive at the result. PEO has 110 classes and 27 properties with a logic expressivity of ALCHQ(D) and incorporates concepts defined in Parasite Life Cycle ontology (PLO) to maximize reuse of existing ontological resources. The new version of PEO (ver 0.2) was released for listing and public use at NCBO’s BioPortal in December 2009. |
|
|
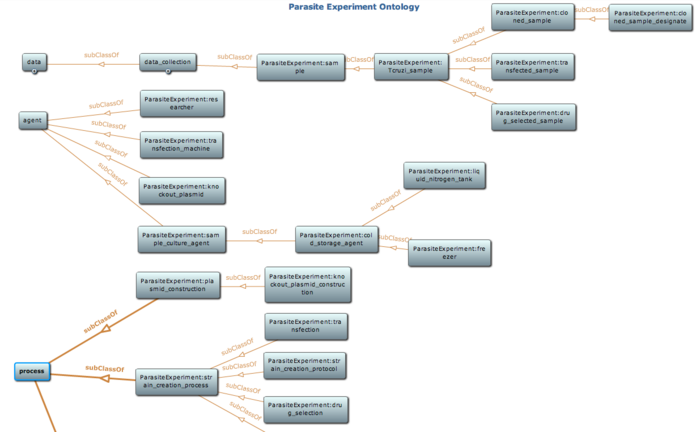 Snapshot of class hierarchy of the parasite experiment ontology using Protege toolkit. Detail of this ontology is available at NCBO BioPortal |
| Alignment of PEO and OBI |
|
These two ontologies present an interesting but different scenario from the OAEI anatomy subtrack. The PEO is being collaboratively developed as part of an NIH funded project to develop and deploy an ontology-driven semantic problem-solving environment for parasite research. The PEO models experiment details and provenance information. It is an application-oriented and more specific domain ontology than OBI, which describes biomedical investigations. In contrast, the OAEI anatomy subtrack ontologies have many common entities since both ontologies describe the same domain (i.e. anatomy) for different species (mouse vs. human). Mapping more specific ontologies such as PEO to a more general ontology such as OBI is an important step towards helping establish a common point of reference that can serve to foster cooperation and interoperability among researchers. Given the explosion of metadata available on the web, our case study to align two related ontologies in the biomedical field has the potential to impact not just the biomedical field but also any research fields that take advantage of semantic web technologies. |
Mappings between PEO and OBI submitted to NCBO BioPortal:
| Feedback/Comments |
|
Please add feedback/comments in Discussion Section on the top of this wiki page. |