PREDOSE
PREDOSE is the acronym for PREscription Drug abuse Online Surveillance and Epidemiology, which is an inter-disciplinary project between the Ohio Center of Excellence in Knowledge-enabled Computing (Kno.e.sis) and the Center for Interventions, Treatment and Addictions Research (CITAR) at Wright State University. The overall aim of PREDOSE is to develop techniques to facilitate prescription drug abuse epidemiology, related to the illicit use of pharmaceutical opioids. PREDOSE is designed to capture the knowledge, attitudes and behaviors of prescription drug abusers through the automatic extraction of semantic information (including entities, relationships, triples and other intelligible constructs such as sentiments, emotions, intervals, frequency, dosage, etc) from social media.
Contents
PREDOSE in the Media
- Researchers use social web forum data to understand nonmedical use of painkillers (article by Wright State University School of Medicine May 6, 2013)
- Semantic App Helps Researchers Understand Prescription Drug Abuse, (article on Semanticweb.com June 11, 2012)
People
Principal Investigators: Raminta Daniulaityte, Amit P. Sheth
Co-Investigators: Robert Carlson, Russel Falck
Graduate Students: Delroy Cameron, Lu Chen, Gary A. Smith, Gaurish Anand, Revathy Krishnamurthy, Nishita Jaykumar, Swapnil Soni
External Collaborators: Drashti Dave, Pablo N. Mendes
Past Members: Kera Z. Watkins , Matthan Sink, Michael Cooney, Sujan Perera, Mandeep Singh, Pratik Desai, Mary Oberer, Kaustav Saha
Overview
The non-medical use of pharmaceutical opioids has been identified as one of the fastest growing forms of drug abuse in the U.S. The White House Office of National Drug Control Policy (ONDCP) has recently launched the Epidemic: Responding to America’s Prescription Drug Abuse Crisis initiative to curb prescription drug abuse problem, mainly through education and drug monitoring programs. The White House Initiative has been prompted by recent research which associate the rise in prescription drug abuse with two important phenomena: 1) expanded the pathways to heroin addiction and 2) escalating rates of accidental overdose deaths. To combat these trends, public health professionals require timely and reliable information on new and emerging drug trends on prescription drug abuse.
Although existing epidemiological data systems provide critically important information about drug abuse trends, they are often time-lagged. Hence, there is a critical need for epidemiological sources that could complement existing drug trend monitoring systems and enhance their capacity for early identification of new and emerging trends. The World Wide Web (Web) has been identified as one of the leading data sources for detecting patterns and changes in the non-medical use of pharmaceutical and other illicit drugs. Many Web 2.0 empowered social platforms, including web forums, provide venues for individuals to freely share their experiences, post questions, and offer comments about different drugs. The PREDOSE project aims to leverage web forum data to provide such timely emerging information on the non-medical use of pharmaceutical opioids.
The PREDOSE project therefore has two(2) specific aims:
| Goals |
|
| Research Problem |
|
Historically, qualitative research in drug abuse interventions programs have relied on manual data collection practices. Data have been gathered from interactive interview sessions with single or groups of users. Interviews are typically transcribed into text, then manually annotated by researchers, to identify themes from the interview sessions. This process is called qualitative coding. Qualitative research tools such as NVivo, have been use to facilitate such Content Analysis. However, the intensive manual effort required to perform the annotations is not scalable and hence, is not practical for Web-based data. Instead, to effectively process the large volume of heterogeneous Web-based data, the field requires a highly automated way of collecting, processing and analyzing semantic metadata from the web. |
| Proposed Solution |
|
To automate the extraction of semantic metadata researchers from the Kno.e.sis Center at Wright State University endeavor to build on prior research to address the complex problem In particular, researchers at Kno.e.sis have successfully applied Semantic Web techniques, to account for shortcomings in Machine Learning and Natural Language Processing techniques to automatically extract knowledge from structured biomedical text and social media (specifically tweets). Substantial progress in understanding content and identify social perceptions from informal text from sources including MySpace, Facebook, and Twitter has been made, through metadata extraction and spatio-temporal-thematic analysis (i.e., semantic analysis). These cutting-edge information processing techniques, with appropriate adaptations can now be exploited to fit the needs of public health and drug abuse research on conversational and informal text, such as those occurring in web forums. |
| Research Plan |
|
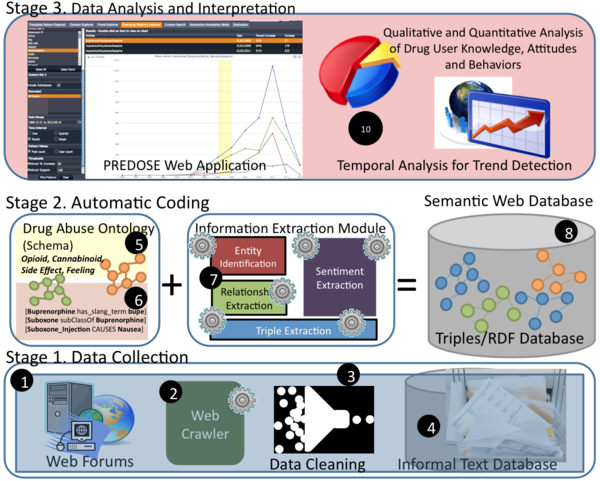
The overall research plan has three(3) distinct stages:
|
Stage 1: Data Collection
- Web Forum Selection: The first component in the PREDOSE platform in stage 1 is for data collection. Web forums selected for the study were chosen based on the following criteria the web forum: 1) allows free discussion of psychoactive drug use; 2) contains information on illicit pharmaceutical drug use, and 3) is publicly accessible. Further, since it is important that this study collects relevant and timely information, such forums are also considered active, both in terms of number of users and diversity in topic discussions.
- Web Crawling: HTML parsers are publicly available to crawl web sites and collect data. Some of these include Nutch, Jericho HTML Parser, HTMLParser etc. In PREDOSE we use the Jericho HTML Parser to write Custom Web Crawlers to crawl data from three online web forums to obtain data for analysis.
- Data Cleaning: We sanitize the crawled HTML and decode special characters in a data cleaning phase that occurs throughout our application where necessary.
- Informal Text Database: Crawled data is stored in a MySQL database together with an index for fast retrieval. We mainly store semantic metadata in the database, based on our information extraction techniques.
Stage 2: Automatic Qualitative Coding
This is the most challenging aspect of PREDOSE. The aim is to use various information extraction techniques to extraction semantic information considered semantically equivalent to qualitative codes, from web forums. Types of extracted information include:
- Drug Abuse Ontology (DAO): We manually created a Drug Abuse Ontology (DAO) to model the prescription drug abuse domain, which is the first ontology on drug abuse in the literature. The current DAO is available online. The DAO is used to facilitate search, and it also serves as the annotation scheme for entity, relationship and triple extraction.
- Entity Identification: from web forum data is challenging because web forums discussions are informal in nature. In particular, web forum data is characterized by a proliferation of slang term references to standard drug references. We leveraged mappings for slang term to known drugs from NIDA, NDCP, Erowid, Urban Dictionary etc to enhance our domain knowledge model. However, while such mappings are a good starting point for entity identification, the more challenging issue of entity disambiguation requires more rigorous techniques. Entity disambiguation is necessary in three scenarios: 1) standard dictionary word disagbiguation (e.g. girl as Gender or the drug Cocaine); 2) word sense disambiguation (i.e., done as Methadone or the act of being done with a task) and finally 3) concept reference disambiguation (i.e. the term "Oxy" may refer to Oxycontin, Generic Oxycontin, Oxycontin OP or Oxycontin OC). We have used a combination of lexical, linguistics and semantics-based techniques to address entity identification and disambiguation: the results of which are reported in our JBI Journal article.
- Relationship Extraction: We have utilized a lexical and semantics-based technique for relationship identification; the details of which are reported in our JBI Journal article.
- Triple Extraction: Previous work at Kno.e.sis have successfully implemented rule-based and probabilistic approaches to triple extraction (Ramakrishnan C, Mendes P. N. and Thomas C. Mehra P), albeit on structured biomedical literature. In another approach Thomas C and Mehra P, etc have implemented a statistical/probabilistic approach to triple extraction also on structured text. Such techniques are not likely apply to informal web forum text. Hence, we implemented a top-down pattern-based technique for triple extraction that utilizes the DAO and the declarative information extraction framework SystemT and it's implementation language AQL (Annotation Query Language).
- Sentiment Extraction - We use an adaptation of the state-of-the-art sentiment extraction extraction technique developed by Chen et al<ref>Lu Chen, Wenbo Wang, Meenakshi Nagarajan, Shaojun Wang and Amit P. Sheth. Extracting Diverse Sentiment Expressions with Target-dependent Polarity from Twitter. In Proceedings of the 6th International AAAI Conference on Weblogs and Social Media (ICWSM), 2012.</ref> to extraction on-target sentiment expressions from web forum data.
- Template Pattern Identification - We use a context-free grammar <ref>D. Cameron, A. P. Sheth, N. Jaykumar, G. Anand, K.Thirunarayan, G. A. Smith. A Hybrid Approach to Finding Relevant Social Media Content for Complex Domain Specific Information Needs (under review)</ref> to define the query language of strings interpretable by PREDOSE. This is a necessary task since many of the complex information needs in PREDOSE require a knowledge of ontological concepts as well as concepts not defined in ontologies such as emotion, sentiment, intensity, frequency, dosage intervals etc.
Stage 3: Data Analysis & Interpretation
In PREDOSE, we developed various components for Content Analysis. These components are included in the PREDOSE web application and the web application developed for Knowledge-Aware Search. More specifically, the PREDOSE Web Application contains components for: 1) Content Analysis and 2) spatio-temporal-thematic analysis.
- Template Pattern Explorer This is a pattern-based component for information retrieval from unstructured texts that; 1) leverages background knowledge to identify lexical variants of ontological concepts in text; 2) has the ability to semantically interpret domain specific elements (e.g. dosage, frequency of use etc) not modeled in background knowledge; 3) enables finding associations in text between template classes based on proximity, by specifying template patterns (e.g. DRUG:DOSAGE:SIDEEFFECT)
- Custom (Proximity) Search This component is a flexible lightweight extension of the Template Pattern Explorer that facilitates pattern-based search, using ontological concepts and user-specified keywords in close proximity, configurable at runtime.
- Content Explorer is a broad content exploration and annotation environment for content analysis. The exploration component enables analysis of text content restricted by: 1) ontological concepts; 2) user-specified keywords; 3) specific data sources and 4) user-specified time ranges. The annotation component supports the creation of training data for information extraction tasks such as: 1) entity identification and 2) sentiment extraction ubiquitous to the project.
- Trend Explorer is a component for longitudinal data analysis based on statistical aggregation of ontological concept mentions and sentiment expressions occurring text based on frequency counts and user activity.
- Emerging Patterns Explorer is an extension of the Trend Explorer for trend analysis of concomitantly occurring ontological concepts and user-specified keywords. This component is most significant because the ability to detect spikes in discussions based on frequently co-occurring terms, unbeknownst to researchers.
A detailed description of the PREDOSE platform is available in our recently published paper<ref> D. Cameron, G. A. Smith, R. Daniulaityte, A. P. Sheth, D. Dave, L. Chen, G. Anand, R. Carlson, K. Z. Watkins, R. Falck. PREDOSE: A Semantic Web Platform for Drug Abuse Epidemiology using Social Media Journal of Biomedical Informatics. 2013 (in press)</ref> in the Journal of Biomedical Informatics.
Loperamide-Withdrawal Discovery
In the early stages of the PREDOSE project we made a discovery, now reported in the literature<ref>R. Daniulaityte, R. Carlson, R. Falck, D. Cameron, S. Perera, L. Chen, A. P. Sheth. "I Just Wanted to Tell You That Loperamide WILL WORK": A Web-Based Study of Extra-Medical Use of Loperamide. Journal of Drug and Alcohol Dependence. 130(1-3): 241-244, 2013. ScienceDirect, PMID 23201175</ref> <ref></ref>
Based on the lexical and semantics-based techniques for entity identification we were able to isolate datasets by drugs based on the mapping of slang references to standard concepts. In the dataset on Loperamide (an over the counter drug for treating diarrhea) we observed that users reported taking the anti-diarrhea treatment drug Loperamide (same as Imodium) to self-medicate from withdrawal symptoms.
The opioid addictions treatment drugs Buprenorphine and Methadone are commonly known for treatment of such withdrawal symptoms. Until now, it was unknown that Loperamide, which is a diarrhea treatment drug can be (or is being used) for the same purpose. Whichismore, we observed that users reported the possibility of mild psychoactive (opiated) effects from megadosing; which is the practice of taking severely excessive amounts of a drug.
PREDOSE Live
http://knoesis-hpco.cs.wright.edu/predose/ [Video Demo]
http://knoesis-hpco.cs.wright.edu/knowledge-aware-search [Video Demo]
Publications
<references/>
- D. Cameron, V. Bhagwan, A. P. Sheth, Towards Comprehensive Longitudinal Healthcare Data Capture. In The 1st International Workshop on the role of Semantic Web in Literature-Based Discovery, SWLBD2012 (co-located with the IEEE International Conference on Bioinformatics and Biomedicine, BIBM2012) Philadelphia PA USA, October 4, 2012, p. 241-247.
Related
- Researchers use social web forum data to understand nonmedical use of painkillers
- Semantic App Helps Researchers Understand Prescription Drug Abuse (news article on semanticweb.com)
- PREDOSE @CITAR
- Knowledge-Aware-Search
- U.S. Targeting Prescription Drug Abuse
- Twitter Helps Determine "Morning People" and "Night Owls"
Funding
This project is sponsored by the National Institutes of Health (NIH) Grant No. R21 DA030571-01A1 awarded to the Ohio Center of Excellence in Knowledge-enabled Computing (Kno.e.sis) and the Center for Treatment, Interventions and Addictions Research (CITAR) titled “A Study of Social Web Data on Buprenorphine Abuse using Semantic Web Technology.” Any opinions, findings, conclusions or recommendations expressed in this material are those of the investigator(s) and do not necessarily reflect the views of the National Institutes of Health.
Contact: Delroy Cameron