Difference between revisions of "Knoesis Alchemy of Healthcare"
(Created page with "'''Knoesis Alchemy of Healthcare''' A semantic platform providing the rich functionality of NLP and Machine learning over an integrated knowledge base comprising of diverse do...") |
|||
| Line 33: | Line 33: | ||
“Cloning and characterization of the mouse MColn1 gene reveal an alternatively spliced transcript not seen in humans.” | “Cloning and characterization of the mouse MColn1 gene reveal an alternatively spliced transcript not seen in humans.” | ||
| − | For this particular example, in figure 1 below, we illustrate how our approach extracts relations by employing knowledge source for contextual enrichment of the content. | + | For this particular example, in figure 1 below, we illustrate how our approach extracts relations by employing knowledge source for the contextual enrichment of the content. |
| − | + | [[File:relation extraction.png|700px|center]] | |
===Personalized Health Knowledge Graph (PHKG) from Unstructured Text=== | ===Personalized Health Knowledge Graph (PHKG) from Unstructured Text=== | ||
| − | A “personalized” Health Knowledge Graph (HKG) needs to be created because health conditions (e.g., symptoms, side-effects, prescribed drugs, living conditions) of two patients with the same disease will likely be different. Such insight resource that is specific to the patient, will provide a more meaningful support for clinicians to intervene appropriately. For instance, Patient A and Patient B are suffering from Asthma while Patient A is moderately allergic to Pollen while Patient B is severely affected by Pollen. kAofH provides a platform that can help create PHKG with integrated appropriate knowledge sources. | + | A “personalized” Health Knowledge Graph (HKG) needs to be created because health conditions (e.g., symptoms, side-effects, prescribed drugs, living conditions) of two patients with the same disease will likely be different. Such insight resource that is specific to the patient, will provide a more meaningful support for clinicians to intervene appropriately. For instance, Patient A and Patient B are suffering from Asthma while Patient A is moderately allergic to Pollen while Patient B is severely affected by Pollen. kAofH provides a platform that can help create PHKG with integrated appropriate knowledge sources. The figure below outlines the creation of PHKG medical knowledge sources that we utilize in performing semantic annotation of the content and, thus, generating a patient-specific KG. |
| − | [[File:PHKG.png|700px|center]] | + | [[File:PHKG example.png|700px|center]] |
Revision as of 17:17, 25 July 2018
Knoesis Alchemy of Healthcare A semantic platform providing the rich functionality of NLP and Machine learning over an integrated knowledge base comprising of diverse domain-specific medical/healthcare-related knowledge sources and Kno.e.sis health ontologies.
Contents
Overview
There has been a substantial increase in the need for health-related support systems in medicine and self-diagnosis. Existing support systems are developed using labor-intensive statistical learning processes. Our approach utilizes existing human-curated knowledge bases (KBs) in the medical domain and creates a domain-specific knowledge graph. Some of the commonly used KBs in the medical domain is ICDs (International Classification of Diseases), SNOMED-CT (Systematized Nomenclature of Medicine-Clinical Terms) among many others.
We are developing a comprehensive platform, Kno.e.sis Alchemy of Health (kAofH), that combines extensive health knowledge graph, machine/deep learning and natural language processing (NLP) techniques for extracting features from the biomedical, clinical, and general health-related text. The kAofH aims to perform critical and goal-oriented tasks, namely,
- Intent identification
- Implicit entity recognition
- Entity linking using ezDI biomedical knowledge source
- Relationship extraction utilizing compound entities
- Segment labeling of biomedical unstructured/structured data
- Event extraction, nuance extraction
- Domain-specific sentiment mining in healthcare
- Relevant Specialization Identification for Web-Based Intervention
- Severity identification-classification.
- Build a patient and domain-centric evolving knowledge graph (KG)
These components of the kAofH are created based on prior works on NLP and Semantic Technologies at Kno.e.sis. Furthermore, the kAofH platform is envisioned and designed to evolve and support other social good projects at Kno.e.sis.
Use Cases
The prominent use-cases of kAofH include entity and relation extraction from the unstructured text for an evolving knowledge graph.
Entity Recognition Use Case
Health-related and user-generated text from sources such as social media, forums (e.g., Twitter, Reddit, Patient.info) pose a challenge due to nature of the language being used to implicitly express ideas, opinions, and facts. For example, “his tip of the appendix is inflamed”. An entity to be implicitly recognized in this example is ‘Appendicitis’ from the concept ‘inflammation of appendix’. Identification of such implicit mentions requires the presence of domain knowledge (e.g., PubMed Health, UMLS). It gets even more complicated in the biomedical domain where entities seldom occur as single entities. For example, “An excessive endogenous or exogenous stimulation by estrogen induces adenomatous hyperplasia of the endometrium”. In this sentence, adenomatous hyperplasia and endometrium occur as one compound entity, where they could have been captured as single entities. Appropriate background knowledge resources such as UMLS (Unified Medical Language System) can be leveraged for more complex tasks, such as recognition and validation of compound entities.
Relation Extraction Use Case
Extracting relationships from a text is an important task and is key to many health-related applications, including biomedical knowledge discovery. Prior work has modeled the problem of relation extraction using statistical learning methods. Such methods include Bayesian approaches, Markov Random Fields, Co-occurrence methods, Rule-based methods, Mutual Information measures, etc. The requirement of a large amount of labeled data and failure in identifying complex relations have obstructed the influence of such methods. However, with the support of background knowledge of the relevant domain can enhance the efficiency of the model. We present two examples to state our use-case:
- Considering the following example taken from [5] :
“Patient suffering from congenital heart disease was treated with lasix.”
In this straightforward healthcare text, entities that can be identified using domain knowledge are; congenital health disease (DBpedia or UMLS) and lasix (MedDRA or DBpedia). Latter is the brand name of the drug Furosemide. One can employ the relationBayesian approach ( conditional random fields), or state-of-the-art embedding approaches to identify the relationship between entities, that is treated.
- Considering the following example taken from BioInfer, how to identify the relation between biomedical entities:
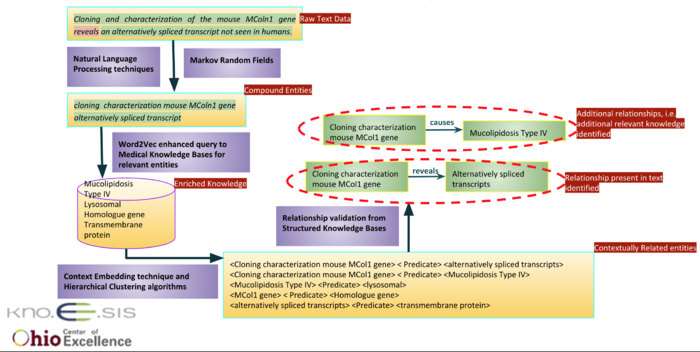
“Cloning and characterization of the mouse MColn1 gene reveal an alternatively spliced transcript not seen in humans.”
For this particular example, in figure 1 below, we illustrate how our approach extracts relations by employing knowledge source for the contextual enrichment of the content.
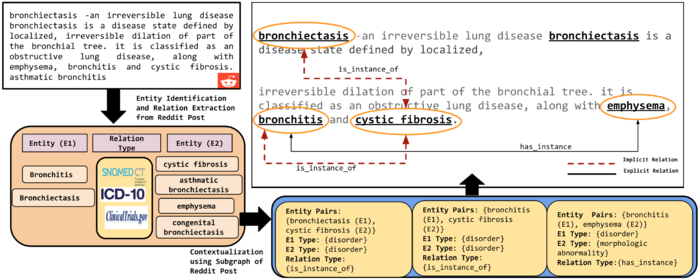
Personalized Health Knowledge Graph (PHKG) from Unstructured Text
A “personalized” Health Knowledge Graph (HKG) needs to be created because health conditions (e.g., symptoms, side-effects, prescribed drugs, living conditions) of two patients with the same disease will likely be different. Such insight resource that is specific to the patient, will provide a more meaningful support for clinicians to intervene appropriately. For instance, Patient A and Patient B are suffering from Asthma while Patient A is moderately allergic to Pollen while Patient B is severely affected by Pollen. kAofH provides a platform that can help create PHKG with integrated appropriate knowledge sources. The figure below outlines the creation of PHKG medical knowledge sources that we utilize in performing semantic annotation of the content and, thus, generating a patient-specific KG.