Biomedical Sciences
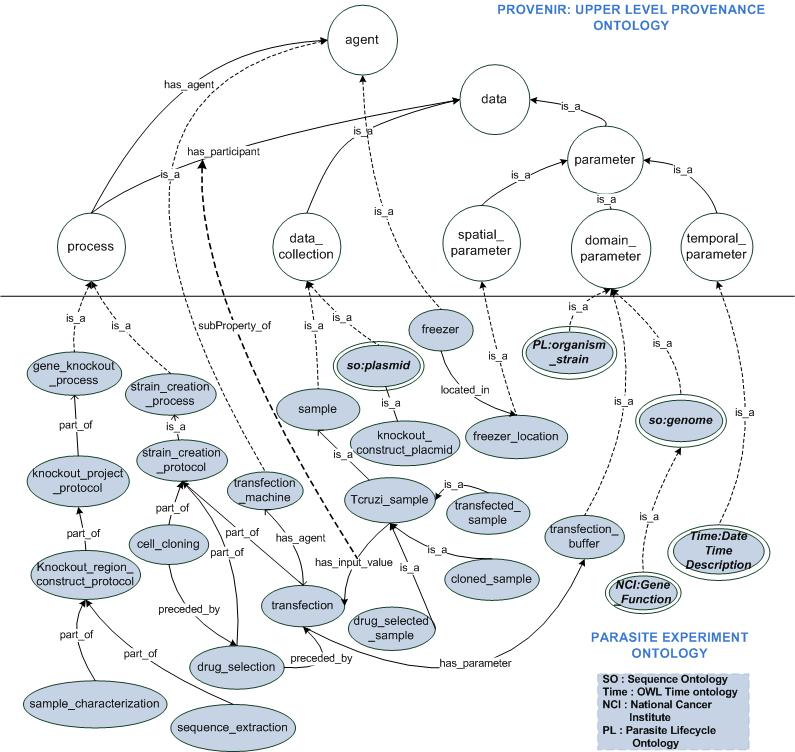
The Provenir Ontology was used as upper level ontology to develop Parasite Experiment Ontology (PEO) for the NIH-funded T.cruzi Semantic Problem Solving Environment (SPSE) project. The PEO extends the classes and relationships in provenir ontology to model provenance information associated with experiment protocols for Gene Knockout (GKO), Strain Creation (SC), Microarray, and Proteome Analysis. These protocols consist of multiple sub-processes, which are modeled in PEO as different classes (Figure 1).
The PEO forms schema to query semantically integrated parasite experimental datasets and databases using intuitive query processing tool, Cuebee. Following are the example provenance queries that can be formulated and successfully executed using PEO and Cuebee:
- List all groups using“target_region_plasmid_Tc00.1047053504033.170_1” target region plasmid.
- Find the name of the researcher who created the knockout plasmid “plasmid66.”
- “cloned_sample66” is not episomal. How many transfection attempts are associated with this sample?
- Which gene was used create the cloned sample “cloned_sample66"?
Reference:
- S.S. Sahoo, D.B. Weatherly, R. Mutharaju, P. Anantharam, A. Sheth, R.L. Tarleton, “Ontology-driven Provenance Management in eScience: an Application in Parasite Research”, The 8th International Conference on Ontologies, DataBases, and Applications of Semantics, (ODBASE 2009), Vilamoura, Algarve-Portugal, Nov 02 - 04, 2009.